Sequence-analysis-pairwise-alignment.pdf
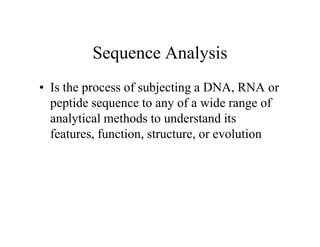
- 1. Sequence Analysis • Is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution • Is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution
- 2. • Given two sequences, we can – Measure their similarity – Determine the residue-residue correspondences – Observe patterns of conservation and variability – Inter evolutionary relationships • Given two sequences, we can – Measure their similarity – Determine the residue-residue correspondences – Observe patterns of conservation and variability – Inter evolutionary relationships
- 3. Bioinformatics Sequence Analysis • The most basic sequence analysis - whether two sequences are related – sequence alignment. This involves aligning two sequences similarity in sequences sequences are related similarity is by chance • The most basic sequence analysis - whether two sequences are related – sequence alignment. This involves aligning two sequences similarity in sequences sequences are related similarity is by chance
- 4. Bioinformatics • Is the most basic tool of bioinformatics. • Sequence similarity must be quantified – important to identify real similarity from coincidence. • Is the most basic tool of bioinformatics. • Sequence similarity must be quantified – important to identify real similarity from coincidence.
- 5. Bioinformatics • Finding similarity between sequences is important for many biological inferences, like •Finding similar proteins allows us to predict the function and structure of the unknown protein. •Similar sequences can come from two species which share a common ancestor indicating their evolutionary relationship. • Locating similar subsequences in DNA allows us to identify pockets of interest, such as regulatory elements.etc • Finding similarity between sequences is important for many biological inferences, like •Finding similar proteins allows us to predict the function and structure of the unknown protein. •Similar sequences can come from two species which share a common ancestor indicating their evolutionary relationship. • Locating similar subsequences in DNA allows us to identify pockets of interest, such as regulatory elements.etc
- 6. Bioinformatics • Pairwise sequence alignment • Local and global alignment • Multiple sequence alignment •Clustal W Sequence Alignment • Pairwise sequence alignment • Local and global alignment • Multiple sequence alignment •Clustal W
- 7. •The comparing of two sequences by searching for a series of individual characters or patterns that are in the same order in the sequences, ie, the identification of residue-residue correspondences. • Local and Global. • Global alignment, attempts to align the entire sequence. If two sequences have approximately the same length and are quite similar, they are suitable for the global alignment. • Local alignment finds stretches of sequences with high level of matches. Pairwise sequence alignment •The comparing of two sequences by searching for a series of individual characters or patterns that are in the same order in the sequences, ie, the identification of residue-residue correspondences. • Local and Global. • Global alignment, attempts to align the entire sequence. If two sequences have approximately the same length and are quite similar, they are suitable for the global alignment. • Local alignment finds stretches of sequences with high level of matches. L G P S S K Q T G K G S - S R I W D N Global alignment L N - I T K S A G K G A I M R L G D A - - - - - - - T G K G - - - - - - - - Local alignment - - - - - - - A G K G - - - - - - - -
- 8. Methods of sequence alignment •Dot plot method • Dynamic programming approach • Smith-Waterman algorithm and Needleman-Wunsch algorithm •Heuristic methods / k-Tuple Method • BLAST and FASTA •Dot plot method • Dynamic programming approach • Smith-Waterman algorithm and Needleman-Wunsch algorithm •Heuristic methods / k-Tuple Method • BLAST and FASTA
- 9. • A dot matrix analysis is a method for comparing two sequences to look for possible alignment (Gibbs and McIntyre 1970) • One sequence (A) is listed across the top of the matrix and the other (B) is listed down the left side • Starting from the first character in B, one moves across the page keeping in the first row and placing a dot in many column where the character in A is the same • The process is continued until all possible comparisons between A and B are made • Any region of similarity is revealed by a diagonal row of dots • Isolated dots not on diagonal represent random matches Dot matrix analysis • A dot matrix analysis is a method for comparing two sequences to look for possible alignment (Gibbs and McIntyre 1970) • One sequence (A) is listed across the top of the matrix and the other (B) is listed down the left side • Starting from the first character in B, one moves across the page keeping in the first row and placing a dot in many column where the character in A is the same • The process is continued until all possible comparisons between A and B are made • Any region of similarity is revealed by a diagonal row of dots • Isolated dots not on diagonal represent random matches
- 10. • Detection of matching regions can be improved by filtering out random matches and this can be achieved by using a sliding window • It means that instead of comparing a single sequence position more positions is compared at the same time and, dot is printed only if a certain minimal number of matches occur • Dot matrix analysis can also be used to find direct and inverted repeats within the sequences Dot matrix analysis • Detection of matching regions can be improved by filtering out random matches and this can be achieved by using a sliding window • It means that instead of comparing a single sequence position more positions is compared at the same time and, dot is printed only if a certain minimal number of matches occur • Dot matrix analysis can also be used to find direct and inverted repeats within the sequences
- 11. • Nucleic Acids Dot Plots - http://arbl.cvmbs.colostate.edu/molkit/dnadot/index.html Dot matrix analysis: two identical sequences
- 12. • Nucleic Acids Dot Plots of genes Adh1 and G6pd in the mouse •http://arbl.cvmbs.colostate.edu/molkit/dnadot/index.html Dot matrix analysis: two very different sequences
- 13. • Nucleic Acids Dot Plots of genes Adh1 from the mouse and rat (25 MY) •http://arbl.cvmbs.colostate.edu/molkit/dnadot/index.html Dot matrix analysis: two similar sequences
- 14. • Nucleic Acids Dot Plots of genes Adh1 from the mouse and rat (25 MY) •http://arbl.cvmbs.colostate.edu/molkit/dnadot/index.html Dot matrix analysis: two similar sequences sequences; size of the sliding window increased
- 15. • Is a highly computationally demanding as well as intensive method. • It aligns two nucleotide/protein sequences, explores all possible alignments and chooses the best alignment (high scoring alignment) as the optimal alignment. • Is based on alignment scores. • It uses gaps to achieve the best alignment. • Global alignment program is based on Needleman-Wunsch algorithm and local alignment on Smith-Waterman. Both algorithms are derivates from the basic dynamic programming algorithm. Dynamic programming algorithm for sequence alignment • Is a highly computationally demanding as well as intensive method. • It aligns two nucleotide/protein sequences, explores all possible alignments and chooses the best alignment (high scoring alignment) as the optimal alignment. • Is based on alignment scores. • It uses gaps to achieve the best alignment. • Global alignment program is based on Needleman-Wunsch algorithm and local alignment on Smith-Waterman. Both algorithms are derivates from the basic dynamic programming algorithm.
- 16. • How are alignments scored? • Using scoring matrices •They account for gaps, substitutions, insertions and deletions. •For nucleic acids, scoring is simple (only 4 characters are present, and substitutions do not happen) •Eg: the scoring scheme used by BioEdit • How are alignments scored? • Using scoring matrices •They account for gaps, substitutions, insertions and deletions. •For nucleic acids, scoring is simple (only 4 characters are present, and substitutions do not happen) •Eg: the scoring scheme used by BioEdit Variation Score Match 2 Mismatch -1 Gap initiation -3 Extending gap by 1 -1
- 17. • For proteins , the scoring schemes are more complicated because amino acid substitutions occur frequently, especially among amino acids with similar physicochemical properties • Eg: Alanine valine substitutions happen without significant changes to the protein.
- 18. Scoring a sequence alignment with a gap penalty Sequence 1 V D S - C Y Sequence 2 V E S L C Y Score 4 2 4 -11 9 7 Score = sum of amino acid pair scores (26) minus single gap penalty (11) = 15 As two sequences may differ, it is likely to have non-identical amino acids placed in the corresponding positions. In order to optimise the alignment gap(s) may be introduced, which may reflect losses or insertions, which occurred in the past in the sequences. Introduction of gaps causes penalties. Scores gained by each match are not always the same, for instance two rare amino acids will score more than two common.
- 19. Derivation of the dynamic programming algorithm 1. Score of new = Score of previous + Score of new alignment alignment (A) aligned pair V D S - C Y V D S - C Y V E S L C Y V E S L C Y 15 = 8 + 7 2. Score of = Score of previous + Score of new alignment (A) alignment (B) aligned pair V D S - C V D S - C V E S L C V E S L C 8 = -1 + 9 3. Repeat removing aligned pairs until end of alignments is reached 1. Score of new = Score of previous + Score of new alignment alignment (A) aligned pair V D S - C Y V D S - C Y V E S L C Y V E S L C Y 15 = 8 + 7 2. Score of = Score of previous + Score of new alignment (A) alignment (B) aligned pair V D S - C V D S - C V E S L C V E S L C 8 = -1 + 9 3. Repeat removing aligned pairs until end of alignments is reached
- 20. • Consider building this alignment in steps, starting from the initial match (V/V) and then sequentially adding a new pair until the alignment is complete, at each stage choosing a pair from all the possible matches that provides the highest score for the alignment up to that point. • If the full alignment has the highest possible (or optimal) score, then the old alignment from which it was derived (A) by addition of the aligned Y/Y pair must also have been optimal up to that point in the alignment. • In this manner, the alignment can be traced back to the first aligned pair that was also an optimal alignment. • The example, which we have considered, illustrates 3 choices: 1. Match the next character(s) in the following position(s); 2. Match the next character(s) to a gap in the upper sequence; 3. Add a gap in the lower sequence. Description of the dynamic programming algorithm • Consider building this alignment in steps, starting from the initial match (V/V) and then sequentially adding a new pair until the alignment is complete, at each stage choosing a pair from all the possible matches that provides the highest score for the alignment up to that point. • If the full alignment has the highest possible (or optimal) score, then the old alignment from which it was derived (A) by addition of the aligned Y/Y pair must also have been optimal up to that point in the alignment. • In this manner, the alignment can be traced back to the first aligned pair that was also an optimal alignment. • The example, which we have considered, illustrates 3 choices: 1. Match the next character(s) in the following position(s); 2. Match the next character(s) to a gap in the upper sequence; 3. Add a gap in the lower sequence.
- 21. • It is critical to have reasonable scoring schemes accepted by the scientific community for DNA and proteins and for different types of alignments • Matrices for DNA are rather similar as there are only two options purine & pyrimidine and match & mismatch • Proteins are much more complex and the number of option is significant • PAM and BLOSUM matrices are the commonly used scoring matrices for proteins. • They are constructed by analysing the substitution frequencies seen in alignments of known families of proteins. • Identities are assigned high positive scores. Also some amino acids are more abundant than others • Frequently observed substitutions also get positive scores. • Mismatches or matches that are unlikely to have been a result of evolution are given negative scores. Scoring matrices • It is critical to have reasonable scoring schemes accepted by the scientific community for DNA and proteins and for different types of alignments • Matrices for DNA are rather similar as there are only two options purine & pyrimidine and match & mismatch • Proteins are much more complex and the number of option is significant • PAM and BLOSUM matrices are the commonly used scoring matrices for proteins. • They are constructed by analysing the substitution frequencies seen in alignments of known families of proteins. • Identities are assigned high positive scores. Also some amino acids are more abundant than others • Frequently observed substitutions also get positive scores. • Mismatches or matches that are unlikely to have been a result of evolution are given negative scores.
- 22. • These scores form the matrix entries and are represented in log odds scores • Odds score is the ratio of chance of amino acid substitution due to essential biological reason to the chance of random substitution. • PAM- (Point Accepted Mutation) matrix is derived from global alignments of very similar sequences, so that an observed change will reflect one mutation • An accepted point mutation is a replacement of one A.A by another, accepted by natural selection • There are many different PAMs, which represent different evolutionary scenarios. • BLOSUM (blocks substitution matrix ) –dvpd from regions of closely related proteins that can be aligned without gaps. They calculated the ratio of observed pairs at any position to the number expected from overall amino acid frequency. • Results in the form of log odds score. • PAM is more suitable for studying quite distant proteins, BLOSUM is for more conserved proteins of domains Scoring matrices • These scores form the matrix entries and are represented in log odds scores • Odds score is the ratio of chance of amino acid substitution due to essential biological reason to the chance of random substitution. • PAM- (Point Accepted Mutation) matrix is derived from global alignments of very similar sequences, so that an observed change will reflect one mutation • An accepted point mutation is a replacement of one A.A by another, accepted by natural selection • There are many different PAMs, which represent different evolutionary scenarios. • BLOSUM (blocks substitution matrix ) –dvpd from regions of closely related proteins that can be aligned without gaps. They calculated the ratio of observed pairs at any position to the number expected from overall amino acid frequency. • Results in the form of log odds score. • PAM is more suitable for studying quite distant proteins, BLOSUM is for more conserved proteins of domains
- 23. • Gap penalties are subtracted from alignment scores to ensure algorithms produce biologically sensible alignments without too many gaps • Gap penalties may be: • Constant – independent of the length of the gap • Proportional – proportional to the length of the gap • Affine – containing gap opening and gap extension contributions. • Opening a gap should be strongly penalised than extending a gap. Gap Penalty • Gap penalties are subtracted from alignment scores to ensure algorithms produce biologically sensible alignments without too many gaps • Gap penalties may be: • Constant – independent of the length of the gap • Proportional – proportional to the length of the gap • Affine – containing gap opening and gap extension contributions. • Opening a gap should be strongly penalised than extending a gap.
- 24. Scoring matrices: PAM (Percent Accepted Mutation) Amino acids are grouped according to the chemistry of the side group: (C) sulfhydryl, (STPAG)-small hydrophilic, (NDEQ) acid, acid amide and hydrophilic, (HRK) basic, (MILV) small hydrophobic, and (FYW) aromatic. Log odds values: +10 means that ancestor probability is greater, 0 means that the probability are equal, -4 means that the change is random. Thus the probability of alignment YY/YY is 10+10=20, whereas YY/TP is –3-5=-8, a rare and unexpected between homologous sequences.
- 25. Scoring matrices: BLOSUM62 (BLOcks amino acid SUbstitution Matrices) Ideology of BLOSUM is similar but it is calculated from a very different and much larger set of proteins, which are much more similar and create blocks of proteins with a similar pattern
- 26. Alignment A: a1 a2 a3 a4 b1 b2 b3 b4 Alignment B: a1 a2 a3 a4 - b1 - b2 b3 b4 Alignment A: a1 a2 a3 a4 b1 b2 b3 b4 Alignment B: a1 a2 a3 a4 - b1 - b2 b3 b4 The highest scoring matrix position is located (in this case s44) and then traced back as far as possible, generating the path shown